Of all the crafty things that metastatic prostate cancer does, this is among the trickiest: it hijacks normal processes that help an embryo grow to become a healthy baby – pathways and programs that nature meant for good – and twists them for its own evil purposes.
Good news: An international team of scientists, funded in part by PCF, has figured out exactly how prostate cancer is turning back the clock. At last, we’ve got proof! “Scientists have suspected for decades that metastatic prostate cancer was using these developmental processes, but it was very challenging to study. Using new methods of examining biopsy specimens, we were able to show this in actual patients’ tumors,” says PCF Young Investigator Mark Pomerantz, M.D., a medical oncologist at the Dana-Farber Cancer Institute and the Broad Institute of Harvard and the Massachusetts Institute of Technology (MIT). Pomerantz is first author of a paper on this landmark research by scientists in the U.S. and Netherlands, published in Nature Genetics. “While it has long been hypothesized that cancer cells reactivate fetal and embryonic programs, we haven’t had experiments in actual human tissue that demonstrate the where and how in the human genome.”
The where and how, in this case, aren’t with the genes themselves, or the genome. They’re in the epigenome (see Box). This is a word you’re going to be hearing a lot, so let’s take a brief look at epigenetics: What Pomerantz is talking about is not a mutation in a gene such as BRCA2; in fact, straightforward genetic changes like that aren’t nearly as common in prostate cancer as they are in some other forms of cancer. This is because, as Pomerantz and colleagues have just shown, prostate cancer seems to be wired differently.
Spaces Between the Genes
The genome and the epigenome are two different things (recall the piano vs the music roll). “For the past 10 years or so, there’s been great focus, led primarily by PCF, to sequence the prostate cancer genome, the DNA code of prostate cancer,” explains Pomerantz. “It has been extremely informative and has led to multiple breakthroughs. However, one of the lessons that we learned by sequencing the genomes of hundreds of prostate cancer patients is that the genome is not the entire story. We have found many mutations in the prostate genome that tell us quite a bit about how the cancer works, but there are other factors that influence what makes prostate cancer form and progress. Our work has primarily been not in the actual genes, but in the vast, vast portions of the genome between the genes.”
So, is this like a tiny version of deep space, those huge stretches of many light years in between stars? Kind of. “Genes only make up a tiny percentage – 1 to 2 percent – of the entire genome,” Pomerantz continues. “The rest is what scientists refer to as dark matter. When I was in biology in high school, they called it ‘junk DNA.’”
But they were wrong; it’s not junky at all. “It turns out that there’s a lot going on in these large spaces between the genes,” Pomerantz says. “There are proteins and other factors – epigenetic changes – coming in and out of this large, noncoding region, that help regulate which genes get turned on and off. There’s an extensive, vast switchboard in this noncoding space. One of our big discoveries a few years ago, supported by PCF, was that the androgen receptor (AR), which drives prostate cancer, interacts with many of these switches in the noncoding DNA, in a very systematic and consistent way in prostate cancer.” In another groundbreaking paper, Pomerantz and colleagues showed that the switches that the AR turns on “shift systematically from normal prostate tissue to cancerous prostate tissue.”
In this latest research, the team focused on finding out what happens in the transition to metastatic prostate cancer. “We observed that the AR makes yet another major transition in where it binds, and the switches it turns on, when we transition from tumor within the prostate to metastatic cancer, and to much more advanced and aggressive cancer. These new switches that are unique to the metastases, that get turned on in the metastases, are sites that are similarly turned on in the human fetal prostate.”
In its malevolent, twisted way, what prostate cancer is doing “sort of makes sense,” says Pomerantz. “That’s one of the amazing things that happens during embryogenesis (the development of an embryo): cells are constantly moving around, recharacterizing themselves. They’re very plastic. That’s what needs to happen for the human body to form. The tumor in the prostate accesses the lowest hanging fruit – these prostatic embryonic switches that allow early prostate cells to move around and change, and do all the things that a metastatic cancer needs to do.”
Tumor Cells Use Existing “Switches”
Normally, these switches, or epigenetic markers, that are turned on in the developing fetus, get turned off once the prostate is formed. Interestingly, they stay off even in cancer “that remains in the prostate,” in low-grade or even intermediate prostate cancer that is fairly slow-growing and content to stay where it is, cancer that doesn’t make that jump to hyperdrive, that doesn’t morph to become metastatic.
But – and this is key – these switches are “bookmarked,” Pomerantz notes. “We were able to identify epigenetic footprints,” bookmarked places in the epigenome, “that are poised to be turned back on. Rather than reinventing the wheel, when cancer cells are finding ways to progress and move out of the prostate, they just grab what is easily visible to them.” These bookmarks are epigenetic tags, also called transcription factors. “We discovered a couple of these transcription factors that are integral to the formation of the prostate in the first place.” One of them is HOXB13; another is FOXA1. “Even though the site has been deactivated.” When re-activated, these tagged switches turn cancer from being differentiated – having well-defined borders, behaving in an orderly way – to de-differentiated, becoming more blobby, wild, and dangerous.
This discovery is the epigenetic equivalent of a smoking gun. It’s the hand in the cookie jar. It’s the trail of bread crumbs left by Hansel and Gretel. And this leads us to potential ways to disrupt cancer’s convenient use of these epigenetic tags.
“These results have profound implications for treatment,” says Pomerantz. “The idea of guiding the cancer cell from its immature, de-differentiated state back to its mature form is not a new concept.” In fact, it’s a successful treatment strategy in certain forms of leukemia. “By identifying the epigenetic programs underlying de-differentiation in prostate cancer, we provide a set of regulatory switches to try to re-differentiate a solid tumor. There are a number of companies that are working on targeting transcription factors, the types of proteins that are being used here.”
One strategy for treatment, then, is to turn back the clock, “to see if we can re-differentiate these cells that are becoming more and more primitive.” It may be, Pomerantz adds, “as we discover more and more of these transcription factors that are bookmarking the sites, and as we find the epigenetic sites that are critically important to drive metastasis, we can begin to attack the factors and the pathways that lead to the production of those factors.” Another strategy for the future may be to somehow un-tag the bookmarks – to make those Hansel and Gretel breadcrumbs disappear, so cancer can’t find them.
This work would not have been possible without “collaboration and input from members of the PCF community around the world,” Pomerantz says. “The co-author list includes many current and former PCF Young Investigators, as well as current PIs. It is a Prostate Cancer Encode project. We have generated the world’s largest prostate epigenomic data set,” which will be the foundation for many future studies. “We have many studies lined up: one is to map not just the AR, but some of these other important factors that work with the AR genome-wide, almost like an epigenetic version of the human genome project. Similar to mapping the genomes of multiple prostates, we will map the epigenome across multiple transcription factors.”
We are confident that we are opening a new vista of investigation and opportunity for intervention,” Pomerantz continues. “The sequencing of the genome got us very far, but not all the way there. By introducing this next ‘ome,’ we are hopeful that we will be able to usher in new approaches to treatment.”
Decades ago, the distinguished British developmental biologist and pioneer in epigenetics, Conrad Hal Waddington, described epigenetics as a contoured valley, with a series of ridges and tracks a cell can follow, like a ball rolling downhill, on its way to being developed (see picture). Pomerantz, building on this model, says that in cancer, instead of gravity and chance taking the ball a certain way, “it’s the ball somehow rolling uphill. The ramp itself is the epigenome, and the ball is the cell, and the ball is made up of the genetic code. But it’s the ramp of the epigenome that takes the cells to their destination.”
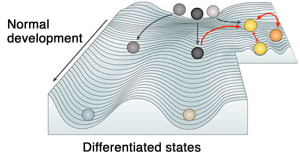
(adapted from Feinberg et al. Nat Rev Genet 2016)
“Every cancer has an altered epigenome,” says molecular biologist and medical oncologist Jonathan Simons, M.D., CEO of the PCF. “Every one. There cannot be a cancer without the epigenomic changes to reprogram the normal genes to abnormal functions, and now we have it comprehensively dissected for advanced prostate cancer.”
PCF was the key to making this research happen, Pomerantz says. “This all began in 2014, when PCF provided the seed funding for us to overcome some of the technical hurdles to map these epigenomic changes. That was absolutely critical. Once we were able to actually explore some of these biological hypotheses, PCF came through again, helping fund these experiments. But just as critical to getting the project off the ground was the opportunity to work with and confer with the rest of the PCF scientific community. The most important meetings were at our PCF scientific retreats, where we sat down with our colleagues to bounce ideas off them and to share resources. Much of the actual experimental material came from collaborators around the world who touched base with us at the PCF meetings.”











